Research Projects
I study the connection between brain structure and function in the brain of mammals. Specifically, I analyze the affect of genes on various brain structures in humans and mice.
Also, I develop new machine learning algorithms and machine vision tools. These tools and algorithms can be applied to genomic data but are not limited to that use.
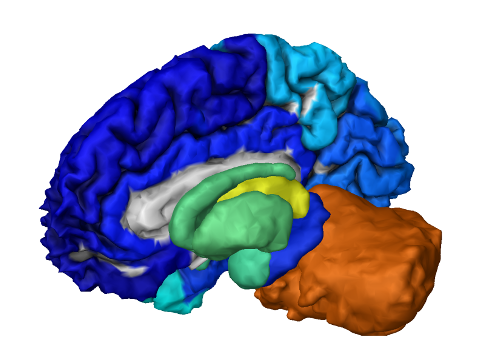
On expression patterns and developmental origin of human brain regions
Genome-wide measurements of gene expression across the human brain can reveal new principles of brain organization and function. To achieve this, we aim to discover which genes are differentially expressed and in what brain regions. We found that almost all genes in the adult human brain bear a developmental ‘footprint’ which determines their areal expression-pattern based on the developmental ontology of brain regions, while at the same time their spatial expression pattern changes during life. Furthermore, pairs of paralog genes with stronger footprint, tend to be more strongly correlated (or anti correlated) suggesting that their expression is more strongly spatially tuned, and these effects are stronger in paralog of synaptic pathways.
Multi-region demixing of brain transcriptome
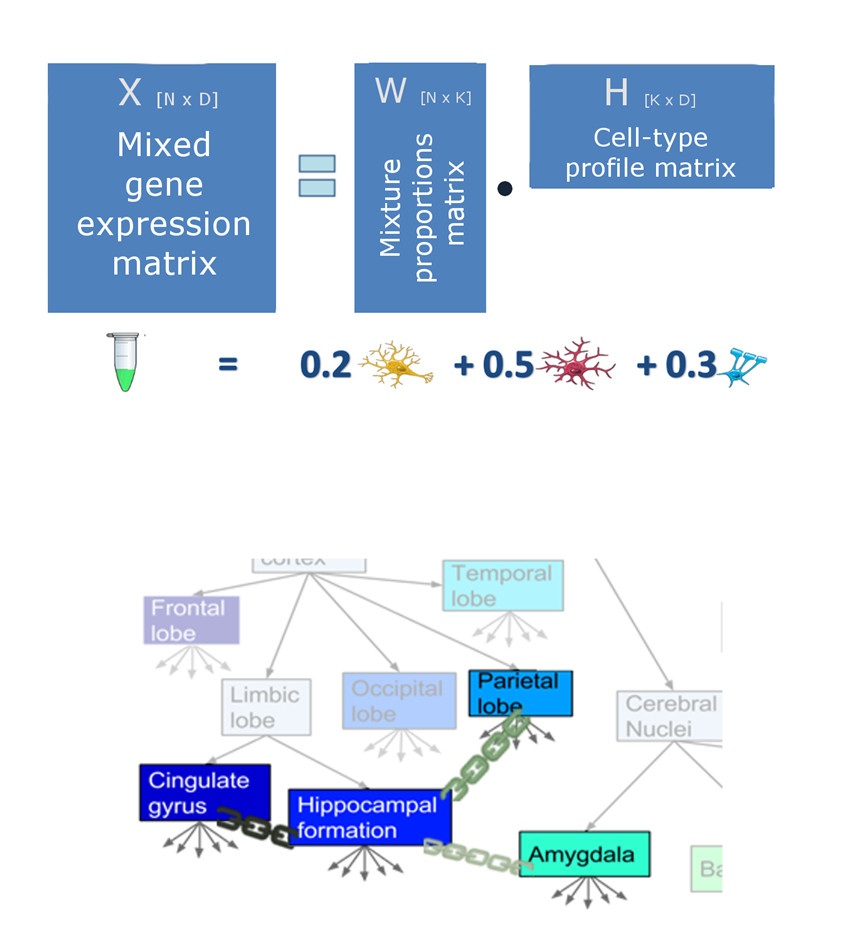
High quality and large scale measurements of gene expression in the brain have been collected across many species and brain regions, but unfortunately, almost all these measurements aggregate over an uncontrolled mix of various cell types including neurons, glia and blood cells. This fact hinders the insights that can be gained by analyzing datasets of gene expression in the brain. Inferring cell-type specific information from these mixtures is a key challenge.
We propose here a probabilistic approach to blindly demix brain transcriptome samples, building on variability of cell-type proportions across samples. We further describe a model that takes into account that tissues in different brain structures differ in their expression profiles by introducing soft-sharing between cell-type-specific profiles from different brain regions. We describe an efficient algorithm to estimate the parameters of the model, as well as techniques to determine some of its hyper parameters from prior knowledge. We study the regimes where multi-region demixing works well using controlled data and evaluate the approach using RNA-seq profiles measured from multiple regions in postmortem human brains. Predictions are validated against ground-truth data of cell-type specific profiles measured in corresponding regions in the mouse brain, showing that soft demixing successfully outperforms baselines of demixing unified and individual regions.
Under review
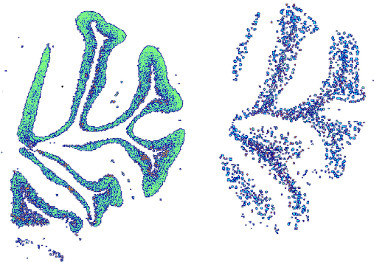
Localizing Genes to Cerebellar Layers by Classifying ISH Images
The way gene expression is spatially distributed across the brain reflects the function and micro-structure of neural tissues. Measuring these patterns is hard because brain tissues are composed of many types of neurons and glia cells, and average gene expression across a region mixes transcripts from many different cells. We present here an approach to identify genes that are primarily expressed in specific brain layers or cell types, based on analyzing high resolution in-situ hybridization images. By learning the spatial patterns of a few known cell markers, we annotate the expression patterns of hundreds of new genes, and predict the layers and cell types they are expressed in.
Here's our paper "Localizing genes to brain layers using ISH image classification" (PLoS Computational Biology, 2012) and its accompanying webpage.
Optimal network structure for spectral radius related phase transitions
The dynamics of contact processes on networks is often determined by the spectral radius of the networks adjacency matrices. A decrease of the spectral radius can prevent the outbreak of an epidemic, or impact the synchronization among systems of coupled oscillators. The spectral radius is thus tightly linked to network dynamics and function. As such, finding the minimal change in network structure necessary to reach the intended spectral radius is important theoretically and practically. Given contemporary big data resources such as large scale communication or social networks, this problem should be solved with a low runtime complexity.
We introduce a novel method for the minimal decrease in weights of edges required to reach a given spectral radius. The problem is formulated as a convex optimization problem, where a global optimum is guaranteed. The method can be easily adjusted to an efficient discrete removal of edges. We introduce a variant of the method which finds optimal decrease with a focus on weights of vertices. The proposed algorithm is exceptionally scalable, solving the problem for real networks of tens of millions of edges in a short time.